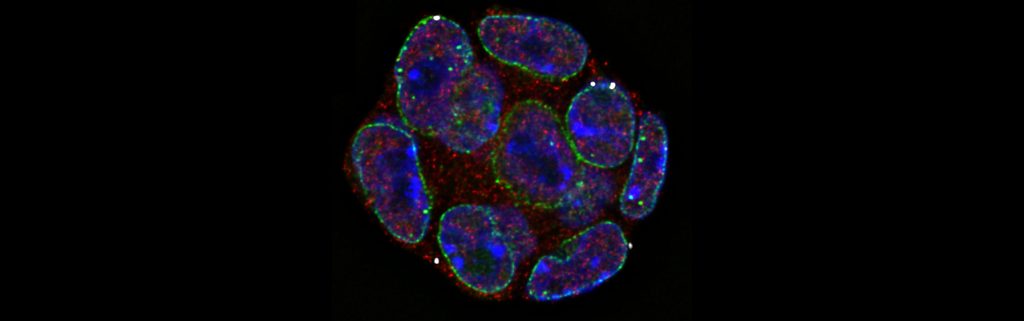
Preterm labour is a global concern, lacking effective treatments due to the poorly understood molecular mechanisms underlying contractions in the uterine muscle tissue known as the myometrium. Gene regulation occurs via a complex interplay between transcriptional factors and regulatory regions that allow the uterus to transition from a quiescent to a contractile phenotype. My research project focuses on the role of transcriptional regulation in the myometrium to better understand the process of pregnancy and labour. For this, I commonly carry out experiments such as qPCR and those involving next generation sequencing techniques, namely ATAC-seq and ChIP-seq, on myometrium tissue.

Gene transcription is controlled in a precise spatial and temporal manner during development and this is known to be controlled by non-coding regulatory elements that can be located at kilobase to megabase distances from the genes they regulate. To properly regulate their cognate genes, enhancers need to be brought into close spatial proximity to them, however, the precise signals directing these long-range chromatin interactions are currently unknown. The goal of my project is to understand at the sequence level, what regulatory code is needed to direct these long-range interactions. To do this I will be using species sequence conservation data to direct CRISPR based chromatin modifications to alter chromatin topography.
PhD Candidates

Trophoblast stem-cells are the first lineage commitment that the early embryo makes after fertilization and go on to become the placenta and extra-embryonic tissues of the fetus. Our collaborators have previously discovered a critical, maternally imprinted gene, Sfmbt2, which has seen to be required for the normal development of fetus by allowing the trophoblast stem-cells to divide indefinitely. My project aims to identify what cis-regulatory elements regulate Sfmbt2 and whether they are also responsible for its parent-of-origin expression. To understand its regulation, I employ CRISPR-Cas9 mediated deletions, gene expression using qPCR, Site-Directed Mutagenesis, enhancer reporter assays, and in vivo studies using genome edited mice.

Enhancers are DNA regulatory elements that control cell-type specific gene expression and are crucial for organism development. Despite their importance, we do not have a good understanding of how this function is encoded into the enhancer DNA sequence. My research focuses on using machine learning, along with comparative genomics and genome evolution to learn the enhancer sequence code.

Embryonic development relies on stem cells and their differentiation into varying cell types, this process works by turning genes on or off in a specific time and space. Since all the cells in the body have an identical genome, there are regulatory elements within the DNA controlling gene expression and therefor cell identity. These elements are termed enhancers, and my project works to further understand these regions by assessing and modifying their sequences, epigenetic landscape, and surrounding chromatin in mouse embryonic stem cells. I have been able to create synthetic regulatory sequences that were confirmed in vitro, and I am now working towards testing these within a genomic context to uncover the regulatory code that enhancers follow. Common experiments I execute in the lab include CRISPR, ChIP-qPCR, cloning, and reporter assays.

The myometrium, a thick muscular layer of the uterine tissue, plays an important role in various reproductive functions along with a key role in labour. There has been remarkable progress towards understanding the physiology and clinical pathophysiology of the myometrium, however, the molecular mechanisms and genetic basis underlying its activity are very poorly understood. Gene regulation is a result of the complex interplay between transcriptional factors and regulatory regions within our DNA which allow the myometrium to transition into various phenotypes. My research project focuses on the role of these myometrium transcriptional factors and their regulation processes during the various stages of pregnancy and labour. For this, I aim to carry out experiments involving next generation sequencing techniques on myometrium tissue and conduct downstream genetic analysis.
Research Associate, Research Communications Officer
Past Lab Members

Ian Tobias
Postdoc

Lida Langroudi
PhD

Gurdeep Singh
PhD

Sakthi Moorthy
Postdoc

Gregor McEdwards

Harry Zhou
Master's

Anandi Bhattacharya
Master's

Mike Schwartz
Master's
Echo Jing

Luis Abatti
PhD

Virlana Shchuka
PhD

Fei Li

Navroop Dhaliwal
PhD

Nakisa Malek-Gilani
Master's

Julie Chih-yu Chen
Master's

Kevin Wang
Master's